VOC Exploration Example
Welcome to the Ultralytics Explorer API notebook. This notebook introduces the resources available for exploring datasets with semantic search, vector search, and SQL queries.
Try yolo explorer (powered by the Explorer API)
Install ultralytics and run yolo explorer in your terminal to run custom queries and semantic search in your browser.
Community Note ⚠️
As of ultralytics>=8.3.10, Ultralytics Explorer support is deprecated. Similar (and expanded) dataset exploration features are available in Ultralytics Platform.
Setup
Install ultralytics and the required dependencies, then check software and hardware.
!uv pip install ultralytics[explorer] openai
yolo checks
Similarity Search
Utilize the power of vector similarity search to find the similar data points in your dataset along with their distance in the embedding space. Simply create an embeddings table for the given dataset-model pair. It is only needed once, and it is reused automatically.
exp = Explorer("VOC.yaml", model="yolo26n.pt")
exp.create_embeddings_table()
Once the embeddings table is built, you can run semantic search in any of the following ways:
- On a given index/list of indices in the dataset, e.g.,
exp.get_similar(idx=[1, 10], limit=10) - On any image/ list of images not in the dataset - exp.get_similar(img=["path/to/img1", "path/to/img2"], limit=10) In case of multiple inputs, the aggregate of their embeddings is used.
You get a pandas DataFrame with the limit number of most similar data points to the input, along with their distance in the embedding space. You can use this dataset to perform further filtering.
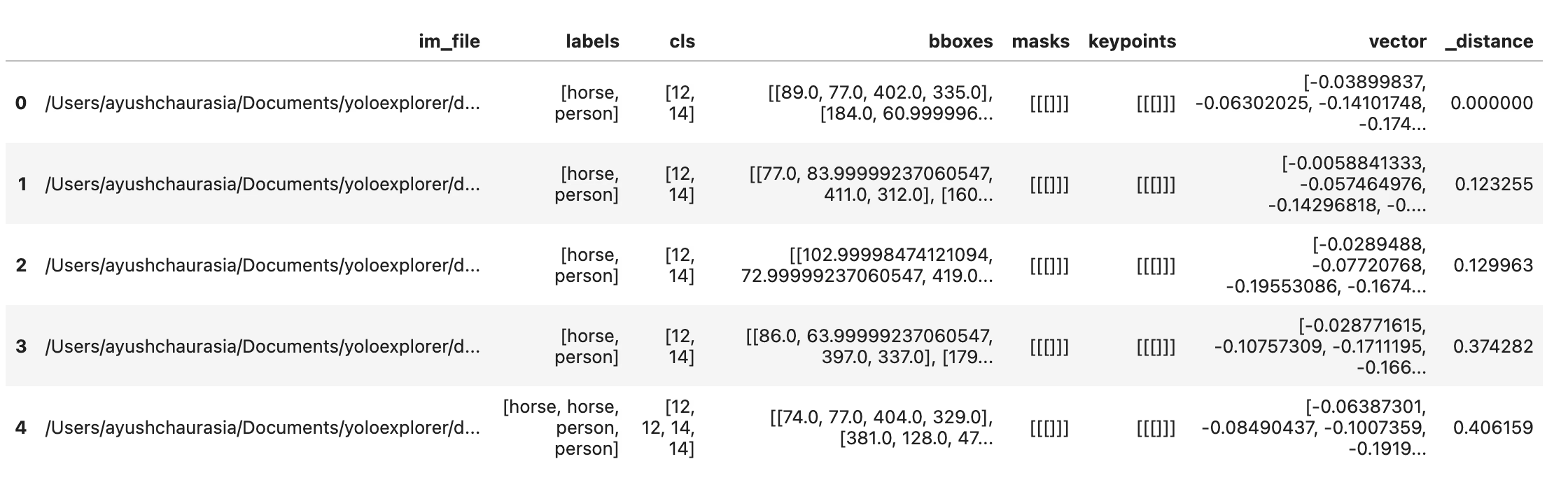
# Search dataset by index
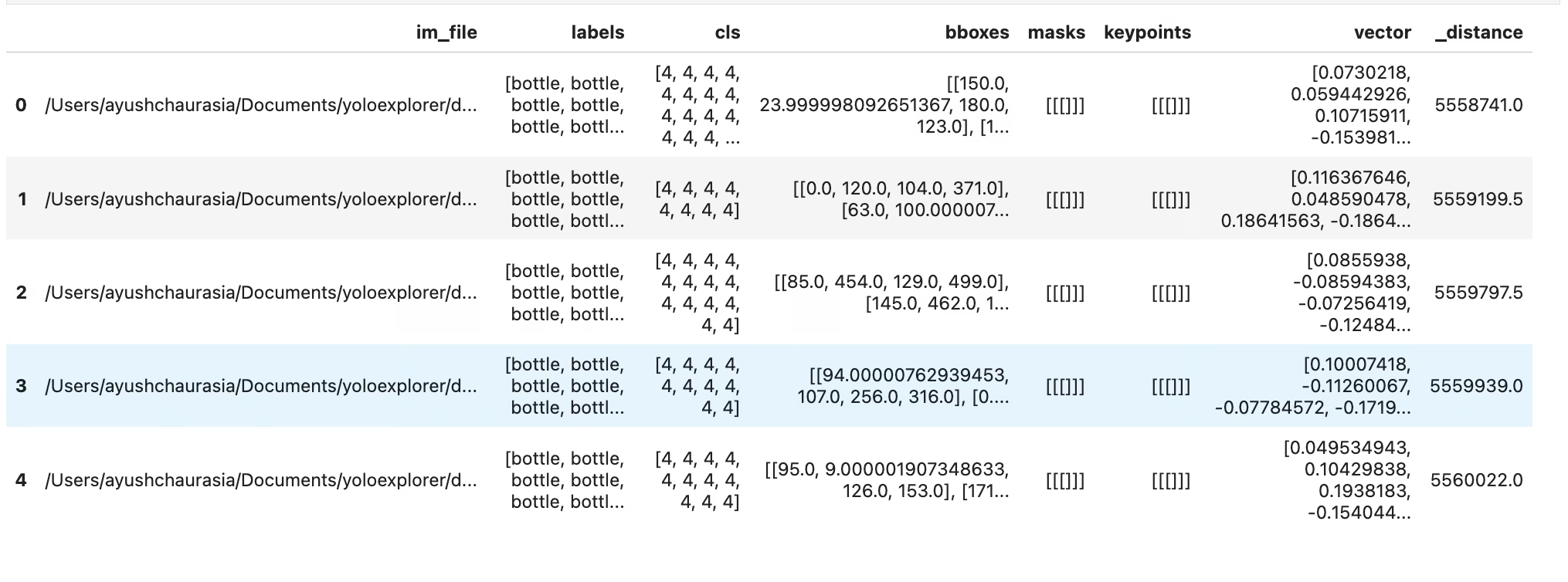
similar = exp.get_similar(idx=1, limit=10)
similar.head()
You can use the also plot the similar samples directly using the plot_similar util
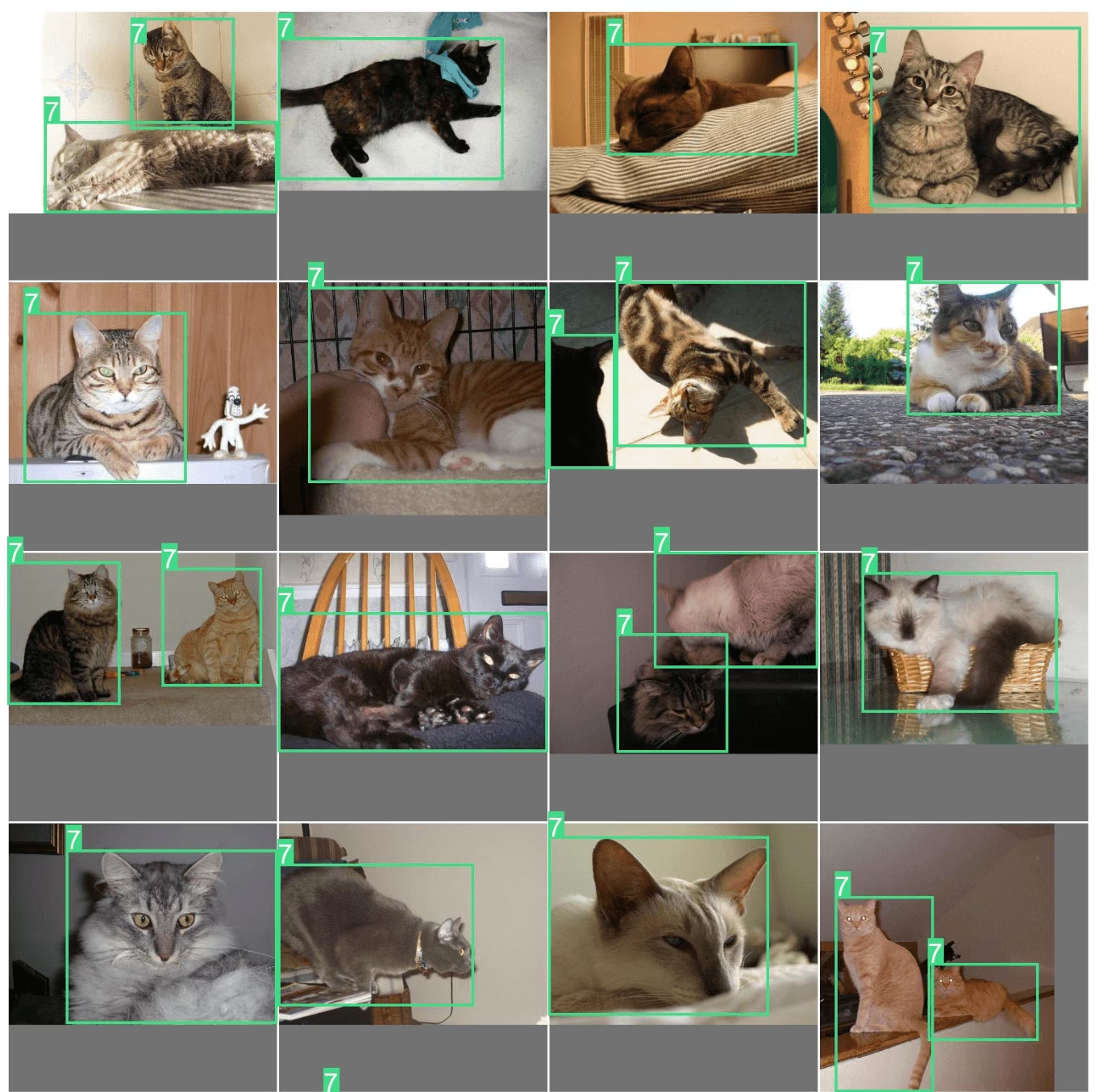
exp.plot_similar(idx=6500, limit=20)
exp.plot_similar(idx=[100, 101], limit=10) # Can also pass list of idxs or imgs
exp.plot_similar(img="https://ultralytics.com/images/bus.jpg", limit=10, labels=False) # Can also pass external images

Ask AI: Search or Filter with Natural Language
You can prompt the Explorer object with the kind of data points you want to see, and it will try to return a DataFrame with those results. Because it is powered by LLMs, it does not always get it right. In that case, it will return None.
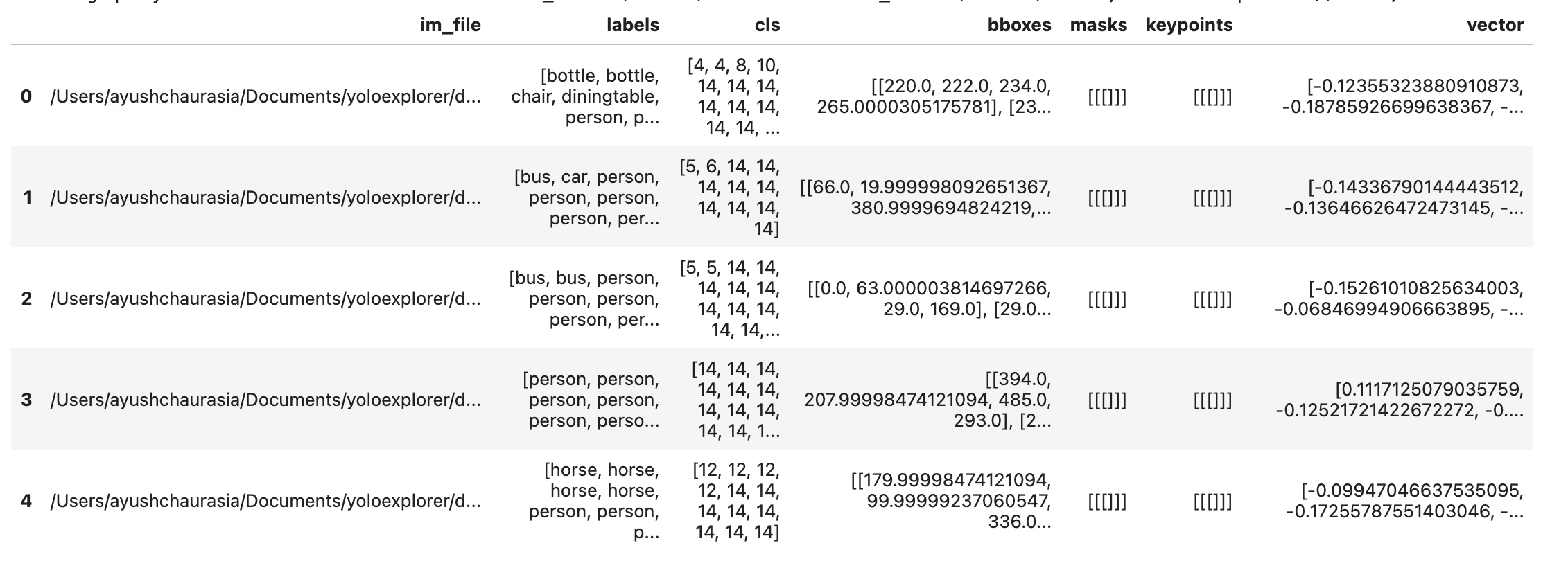
df = exp.ask_ai("show me images containing more than 10 objects with at least 2 persons")
df.head(5)
To plot these results, you can use the plot_query_result utility. Example:
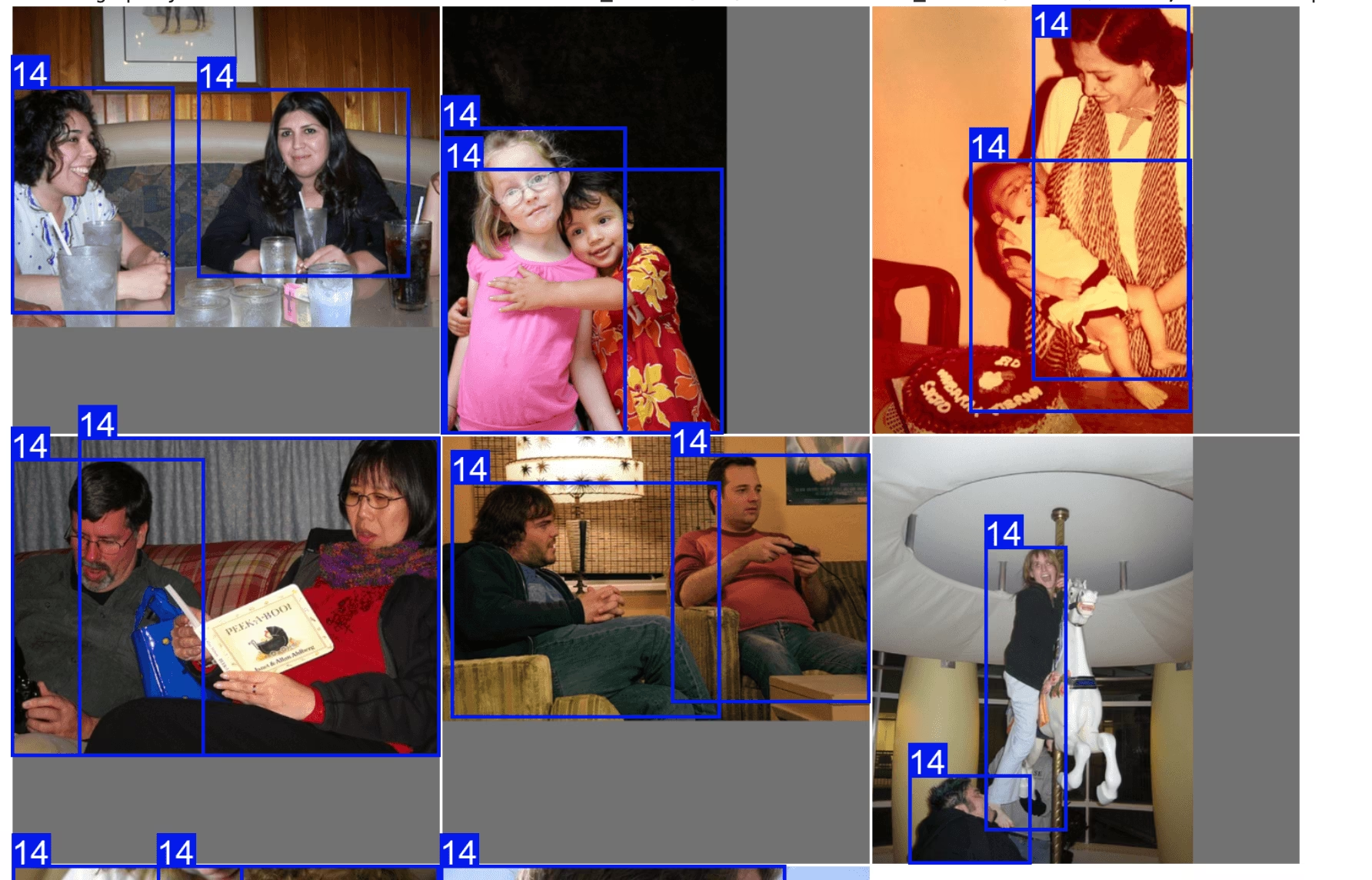
plt = plot_query_result(exp.ask_ai("show me 10 images containing exactly 2 persons"))
Image.fromarray(plt)
# plot
from PIL import Image
from ultralytics.data.explorer import plot_query_result
plt = plot_query_result(exp.ask_ai("show me 10 images containing exactly 2 persons"))
Image.fromarray(plt)
Run SQL Queries on Your Dataset
Sometimes you might want to investigate certain entries in your dataset. For this, Explorer allows you to execute SQL queries. It accepts either of the following formats:
- Queries beginning with "WHERE" will automatically select all columns. This can be thought of as a shorthand query.
- You can also write full queries where you can specify which columns to select.
This can be used to investigate model performance and specific data points. For example:
- let's say your model struggles on images that have humans and dogs. You can write a query like this to select the points that have at least 2 humans AND at least one dog.
You can combine SQL query and semantic search to filter down to specific type of results
table = exp.sql_query("WHERE labels LIKE '%person, person%' AND labels LIKE '%dog%' LIMIT 10")
exp.plot_sql_query("WHERE labels LIKE '%person, person%' AND labels LIKE '%dog%' LIMIT 10", labels=True)
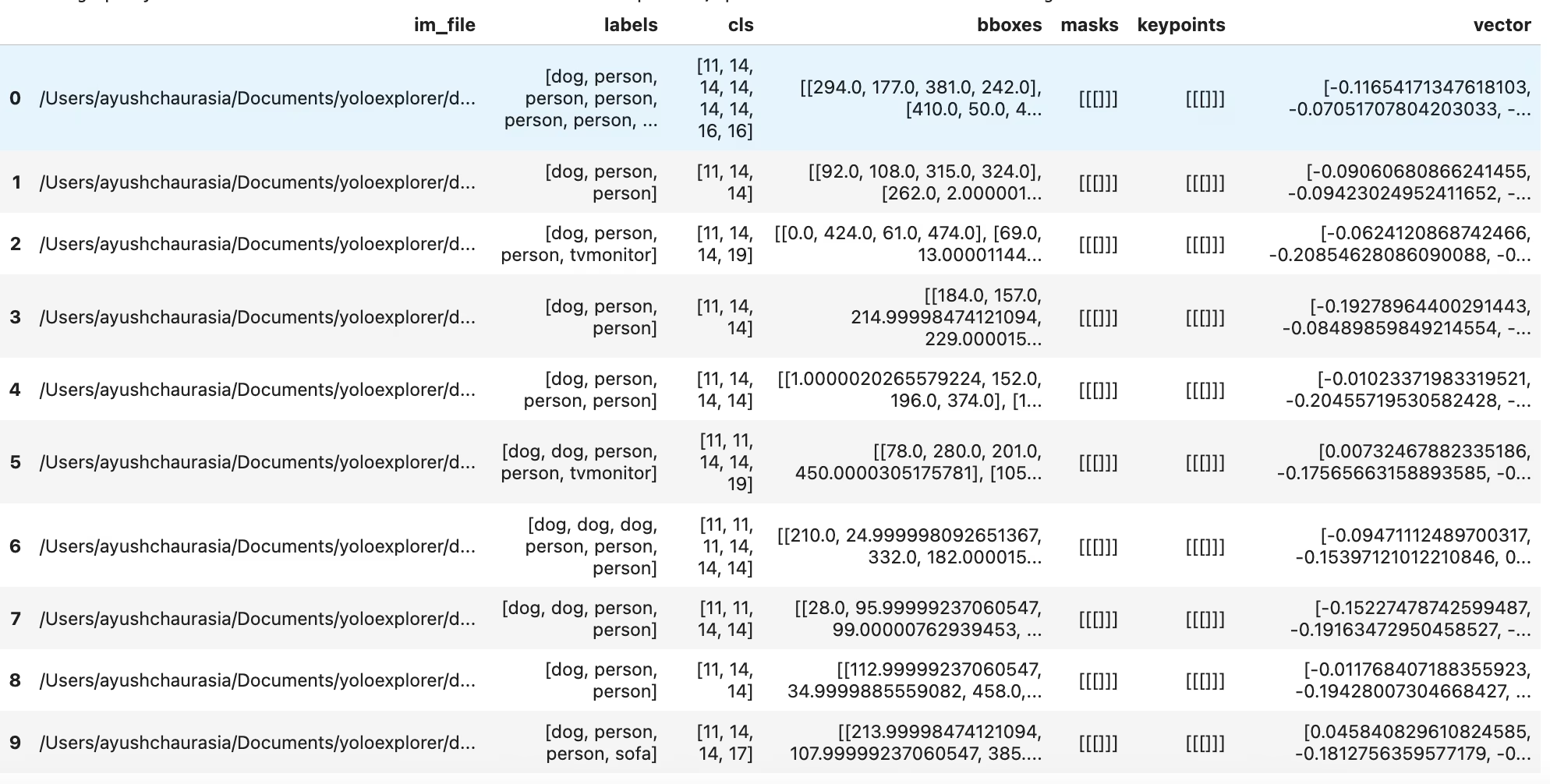
table = exp.sql_query("WHERE labels LIKE '%person, person%' AND labels LIKE '%dog%' LIMIT 10")
print(table)
Just like similarity search, you also get a util to directly plot the sql queries using exp.plot_sql_query
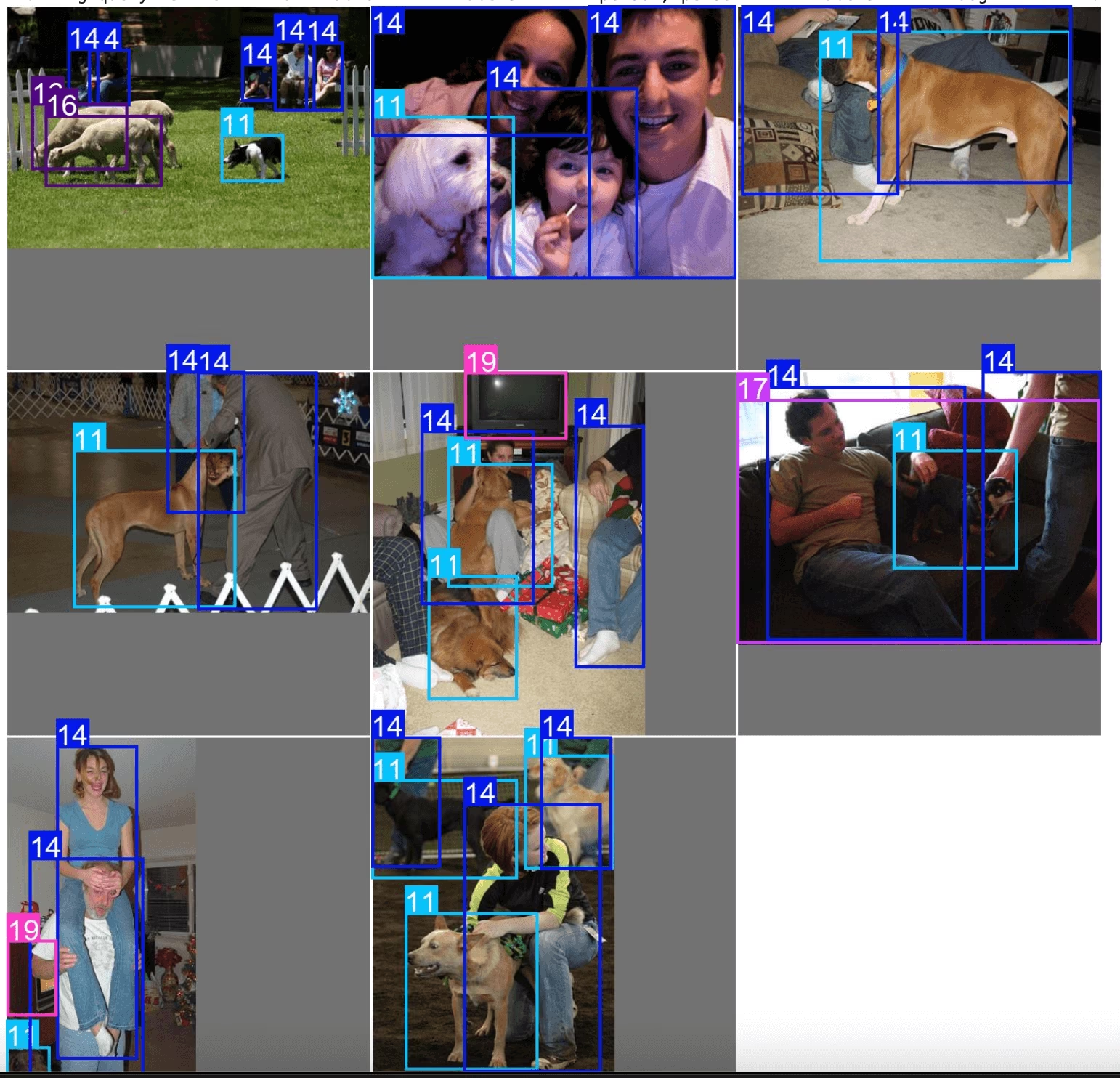
exp.plot_sql_query("WHERE labels LIKE '%person, person%' AND labels LIKE '%dog%' LIMIT 10", labels=True)
Working with embeddings Table (Advanced)
Explorer works on LanceDB tables internally. You can access this table directly, using Explorer.table object and run raw queries, push down pre- and post-filters, etc.
table = exp.table
print(table.schema)
Run raw queries¶
Vector Search finds the nearest vectors from the database. In a recommendation system or search engine, you can find similar products from the one you searched. In LLM and other AI applications, each data point can be presented by the embeddings generated from some models, it returns the most relevant features.
A search in high-dimensional vector space, is to find K-Nearest-Neighbors (KNN) of the query vector.
Metric In LanceDB, a Metric is the way to describe the distance between a pair of vectors. Currently, it supports the following metrics:
- L2
- Cosine
- Dot Explorer's similarity search uses L2 by default. You can run queries on tables directly, or use the lance format to build custom utilities to manage datasets. More details on available LanceDB table ops in the docs
dummy_img_embedding = [i for i in range(256)]
table.search(dummy_img_embedding).limit(5).to_pandas()
Interconversion to popular data formats
df = table.to_pandas()
pa_table = table.to_arrow()
Work with Embeddings
You can access the raw embedding from lancedb Table and analyze it. The image embeddings are stored in column vector
import numpy as np
embeddings = table.to_pandas()["vector"].tolist()
embeddings = np.array(embeddings)
Scatterplot
One of the preliminary steps in analyzing embeddings is by plotting them in 2D space via dimensionality reduction. Let's try an example
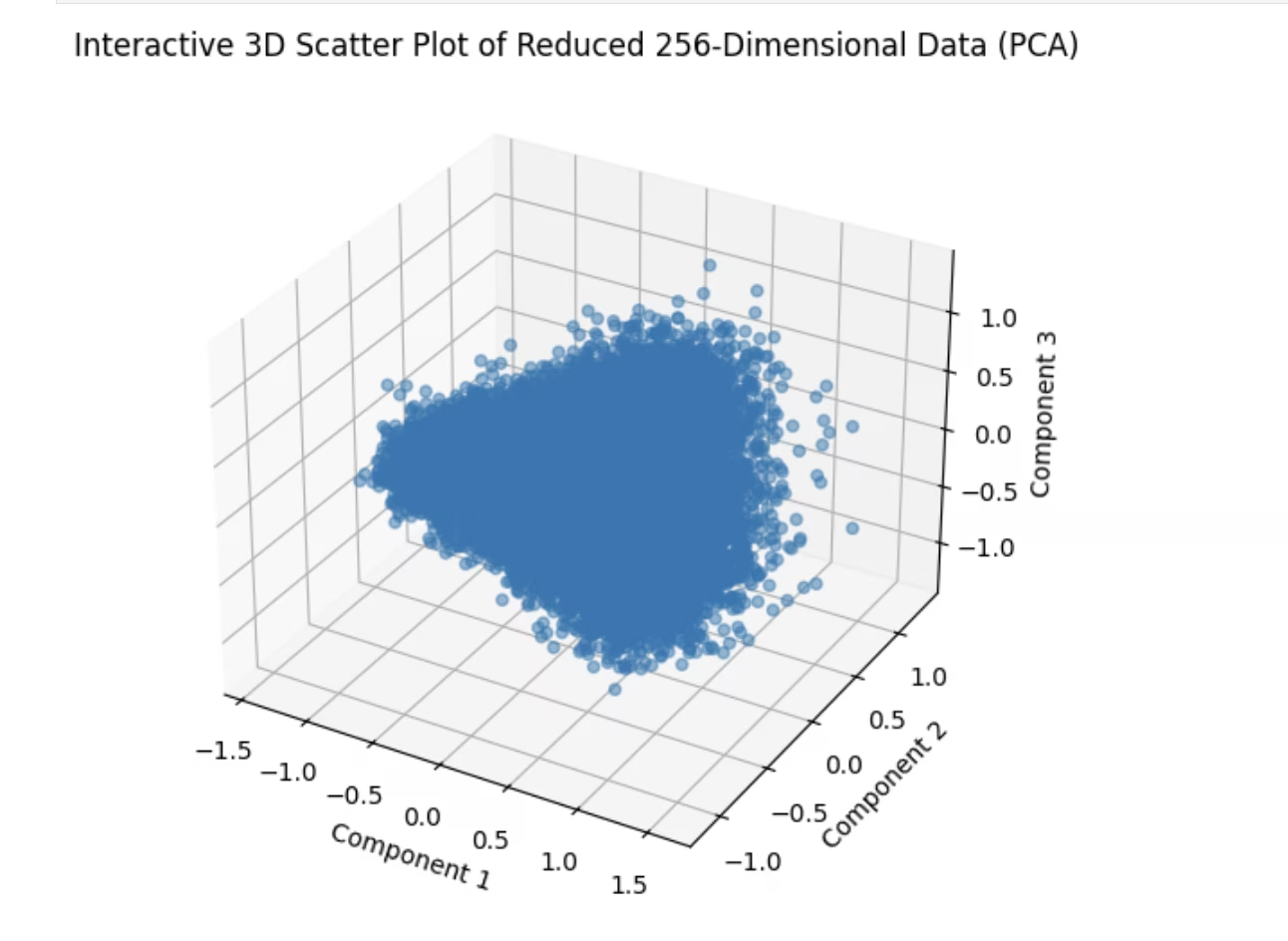
import matplotlib.pyplot as plt
from sklearn.decomposition import PCA # pip install scikit-learn
# Reduce dimensions using PCA to 3 components for visualization in 3D
pca = PCA(n_components=3)
reduced_data = pca.fit_transform(embeddings)
# Create a 3D scatter plot using Matplotlib's Axes3D
fig = plt.figure(figsize=(8, 6))
ax = fig.add_subplot(111, projection="3d")
# Scatter plot
ax.scatter(reduced_data[:, 0], reduced_data[:, 1], reduced_data[:, 2], alpha=0.5)
ax.set_title("3D Scatter Plot of Reduced 256-Dimensional Data (PCA)")
ax.set_xlabel("Component 1")
ax.set_ylabel("Component 2")
ax.set_zlabel("Component 3")
plt.show()
Similarity Index
Here's a simple example of an operation powered by the embeddings table. Explorer comes with a similarity_index operation-
- It tries to estimate how similar each data point is with the rest of the dataset.
- It does that by counting how many image embeddings lie closer than max_dist to the current image in the generated embedding space, considering top_k similar images at a time.
For a given dataset, model, max_dist & top_k the similarity index once generated will be reused. In case, your dataset has changed, or you simply need to regenerate the similarity index, you can pass force=True. Similar to vector and SQL search, this also comes with a util to directly plot it. Let's look
sim_idx = exp.similarity_index(max_dist=0.2, top_k=0.01)
exp.plot_similarity_index(max_dist=0.2, top_k=0.01)
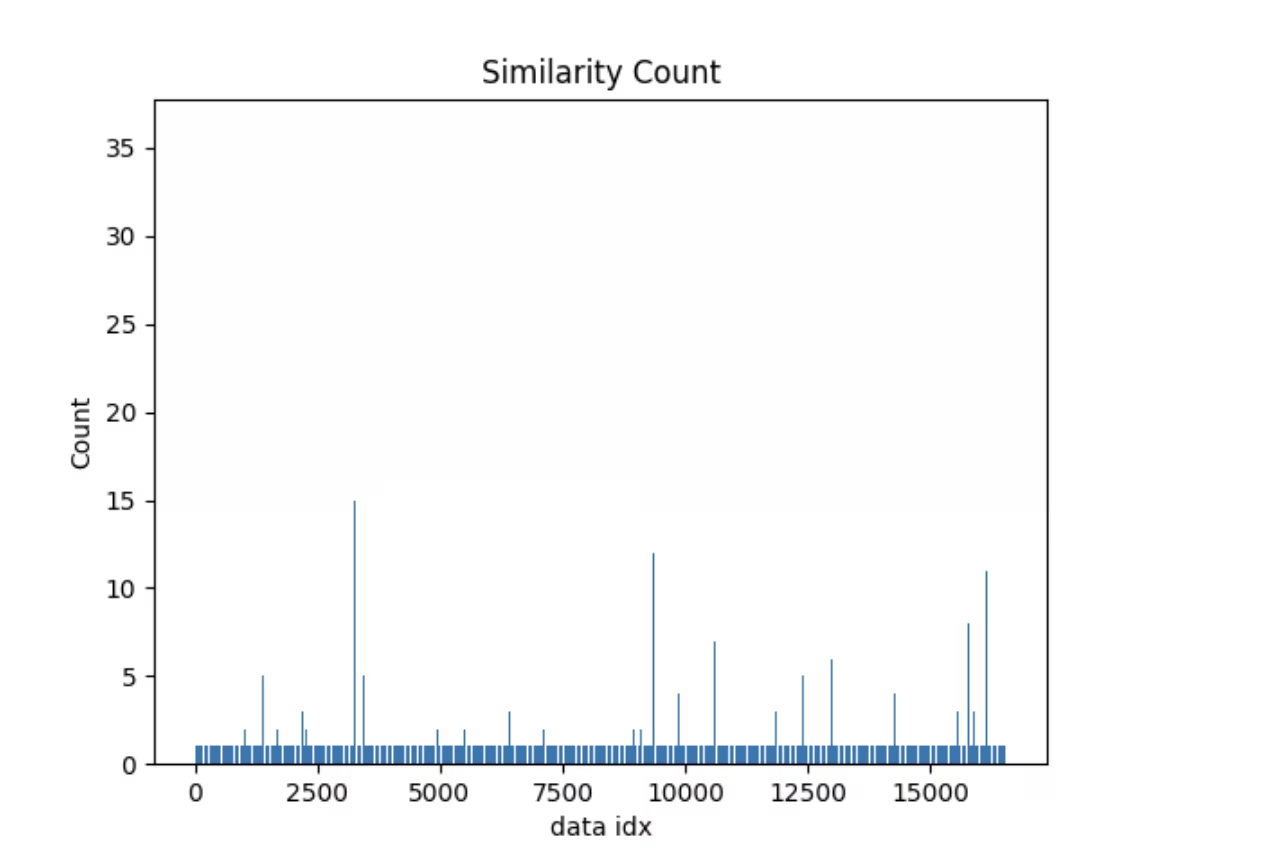
at the plot first
exp.plot_similarity_index(max_dist=0.2, top_k=0.01)
Now let's look at the output of the operation
sim_idx = exp.similarity_index(max_dist=0.2, top_k=0.01, force=False)
sim_idx
Let's create a query to see what data points have similarity count of more than 30 and plot images similar to them.
import numpy as np
sim_count = np.array(sim_idx["count"])
sim_idx["im_file"][sim_count > 30]
You should see something like this

exp.plot_similar(idx=[7146, 14035]) # Using avg embeddings of 2 images